|
|





Abdurrahman Abul-Basher
Research Fellow
Bioinformatics & Applied Machine Learning Scientist

Jump to: News
| Teachings
| Research
Articles
I am a research fellow at Lee Laboratory at Harvard Medical School and Boston Children's Hospital. I work in analyzing cell phenotypic transition dynamics from live-cell images and high-dimensional gene/protien expression data (e.g., transcriptomics, proteomics, and spatial transcriptomics) across human tissues. I am also expert in analyzing large-scale genomics data using cutting edge machine/deep learning algorithms (e.g., contrastive analysis).
I obtained my PhD in Bioinformatics
(an interdisciplinary field) from the University of British Columbia (UBC), where I was advised by Dr. Steven J. Hallam (Hallam Lab). Previously, I obtained my bachelor's in computer
science from King AbdulAziz University (KAU) and masters in
information systems security from Concordia University, where I
worked with Dr. Benjamin C. M.
Fung in the Data Mining and
Security (DMaS) Lab to discover topics from chat logs for crime investigation.
Misc: In my spare time, I enjoy reading novels, history, modern philosophy, and writing poetry.
|
|
|
|
|
Selected fellowships and awards
- Four Year Fellowships (4YF) ($18,200 per year + tuition fee), The University of British
Columbia (UBC), Canada. 2013-2017.
- Faculty of Science - Graduate Support Initiative (GSI) Fund ($8,500 per year), The
University of British Columbia (UBC), Canada.2013-2017.
- Power Corporation of Canada Graduate Fellowships ($5,000), Concordia University, Canada.
2009-2010.
- Concordia Graduate Student Support Program (GSSP) ($15,000 per year), Concordia
University, Canada. 2009-2011.
- First Honor Graduate for graduating with high GPA from King AbdulAziz University, Saudi
Arabia. 2008.
|
|
News
- Pleased to announce that our abstract "Heterogeneity-Preserving Discriminative Feature Selection for Subtype Discovery" got accepted at Cell Symposia: The conceptual power of single cell biology, conference, 2023.
- Pleased to announce that our abstract "Hetero-Discriminative preserving feature selection for subtype discovery" got accepted for Dr. M. Judah Folkman Research Day at Boston Children's Hospital, 2023.
- I gave an invited talk at Boston Children's Hospital, 2022.
- Pleased to announce that our paper Aggregating statistically correlated metabolic pathways into groups to improve prediction performance got accepted at BIOINFORMATICS, 2022.
- Our paper Relabeling metabolic pathway data with groups to improve prediction outcomes got accepted at ICCABS, 2021.
- I gave an invited talk at H. Lee Moffitt Cancer Center & Research Institute, 2021.
- I gave an invited talk at Boston Children's Hospital, Harvard Medical School, Harvard University, 2021.
- Pleased to announce that the source code for leADS
is publicly made available!
- Pleased to hear that our article Metabolic
pathway prediction using non-negative matrix factorization with improved precision
has been accepted at the Journal of computational biology (JCB), 2021!
- I gave an invited talk at Center
for Protein Degradation at Dana-Farber Cancer Institute, Harvard Medical School,
Harvard University, 2021.
- I'm happy to announce that I've received the President’s Academic Excellence
Initiative PhD Award, The University of British Columbia (UBC), 2020!
- I gave a presentation at the 10th International Conference on Computational Advances in
Bio and medical Sciences (ICCABS),
2020.
- Our paper Metabolic
pathway prediction using non-negative matrix factorization with improved precision
got accepted at ICCABS, 2020.
- Our paper Leveraging
heterogeneous network embedding for metabolic pathway prediction got accepted at
Bioinformatics, 2020.
- Our paper Metabolic
pathway inference using multi-label classification with rich pathway features got
accepted at PLOS Computational Biology, 2020.
|
|
|
|
Teachings
|
|
The University of British Columbia, Master of Data Sciences (MDS)
- Graduate Student Instructor, DSCI 571 Supervised Learning I, 2016
- Graduate Student Instructor, DSCI 573 Feature and Model Selection, 2017
- Graduate Student Instructor, DSCI 575 Advanced Machine Learning, 2017
|
|
|
Selected publications
published
accepted
preprint
under preparation
under review
|
|
Heterogeneity-Preserving Discriminative Feature Selection for Subtype Discovery
Abdurrahman Abul-Basher,
Caleb Hallinan,
and
Kwonmoo Lee
Under preparation, 2024
code
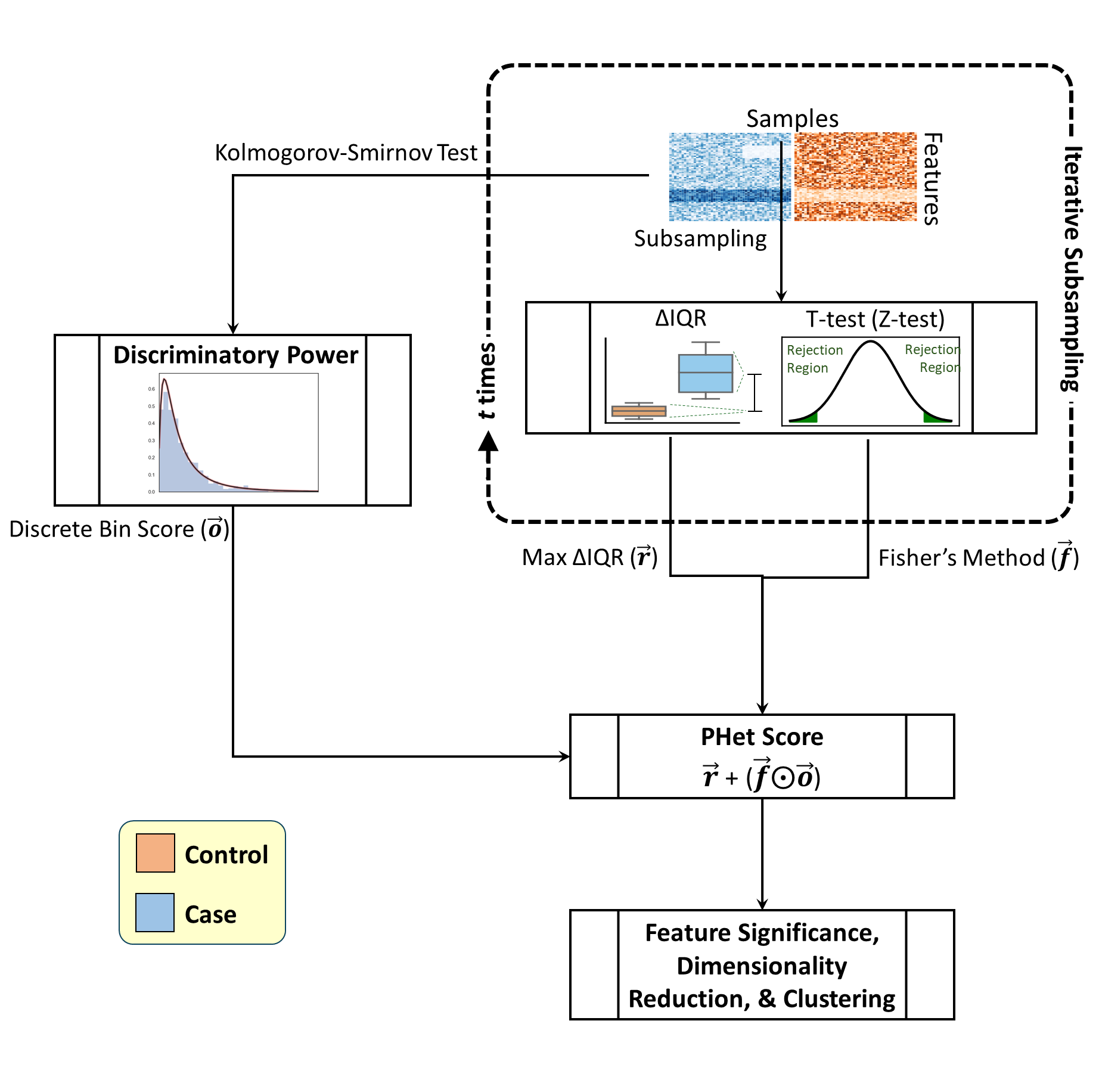
In this article, we present a novel approach, termed PHet (Preserving Heterogeneity), designed to capture the diversity within each disease condition while maintaining the discrimination of known disease states. Our analysis identified features with significant differences in interquartile range (IQR) between classes, indicating crucial subtype information. Validation using public single-cell RNA-seq and microarray datasets demonstrated PHet's effectiveness in preserving sample heterogeneity while maintaining discrimination of known disease/cell states, surpassing the performance of previous outlier-based methods.
|
|
Leveraging multiple (less-trusted) sources to improve metabolic pathway
prediction [TO BE ADDED]
Abdurrahman Abul-Basher,
XXXX
Under preparation, 2024
[TO BE ADDED]
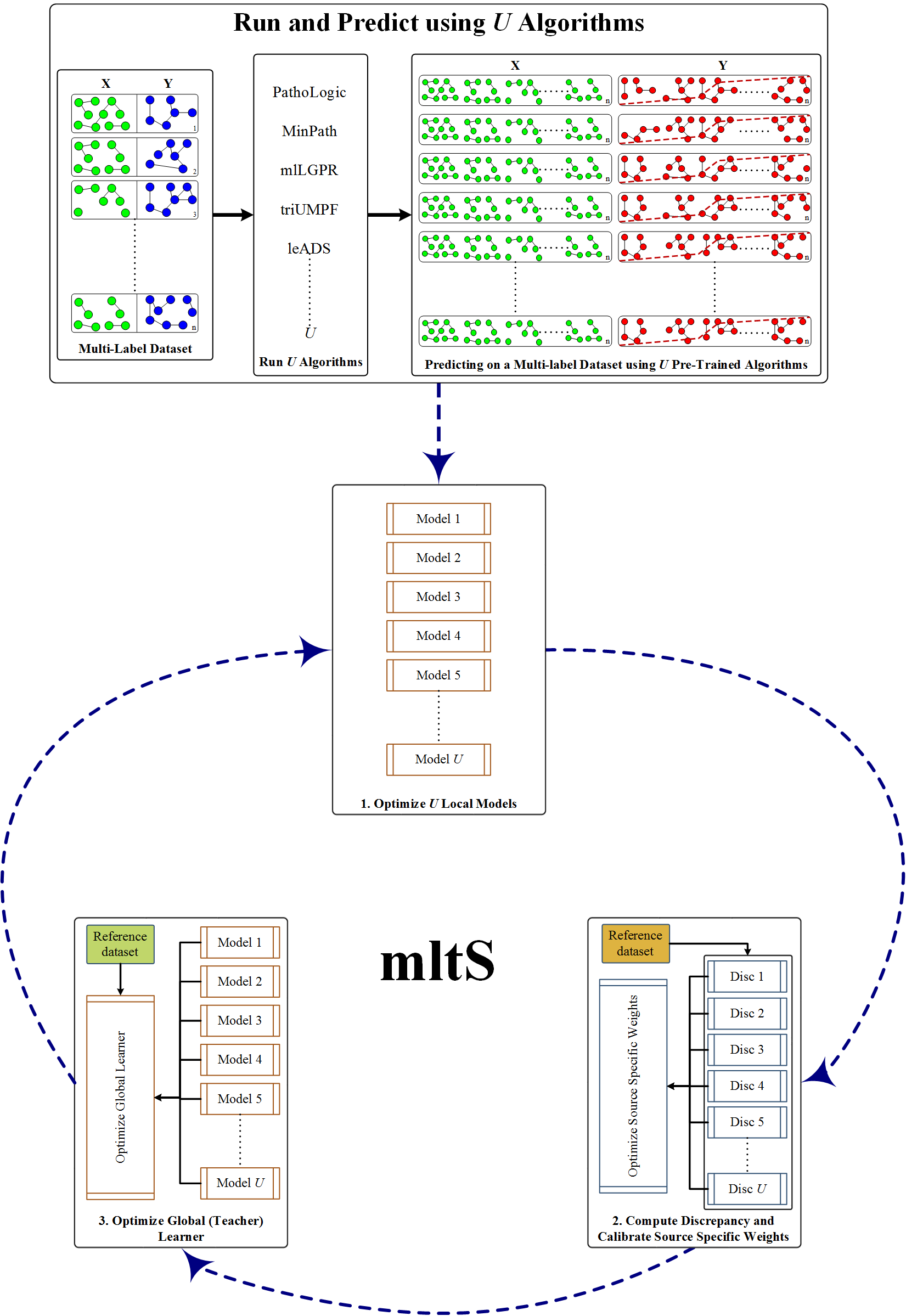
This paper presents mltS (multi-label learning based on less-trusted sources) which is an
ensemble based learning that leverages the idea of estimating memebers reliability scores in
an ensemble given a small reference collection dataset. Using mltS, one can the assess the
reliability of each model while performing inference in three ways: "meta-predict (mp)",
"meta-weight (mw)", and "meta-adaptive (ma)".
|
|
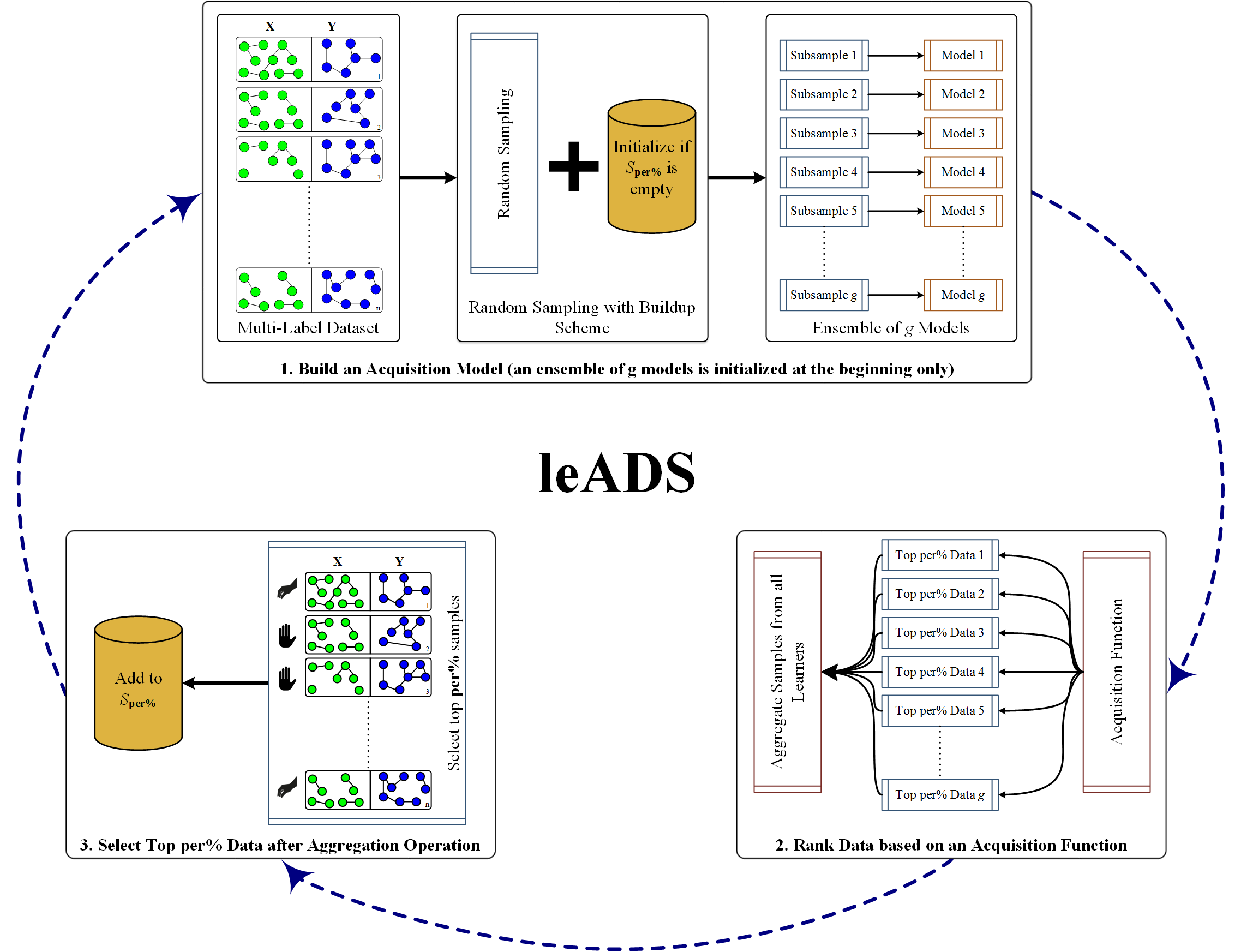
leADS: improved metabolic pathway inference based on active dataset
subsampling
Abdurrahman Abul-Basher,
Aditi N.
Nallan,
Ryan J.
McLaughlin,
Julia Anstett,
and
Steven J. Hallam
Under review, 2024
code
This paper presents leADS (multi-label learning based on active dataset subsampling) that
leverages the idea of subsampling examples from a pool of multi-label data to reduce the
negative impact of training loss.
|
|
Aggregating statistically correlated metabolic pathways into groups to improve prediction performance
Abdurrahman Abul-Basher and
Steven J. Hallam
BIOINFORMATICS (to appear), 2022
code
This paper presents the CHAP (correlated pathway group) package comprising of three hierarchical mixture models: SOAP (sparse correlated pathway group, SPREAT (distributed sparse correlated pathway group), and CTM (correlated topic model) to characterize pathways.
|
|
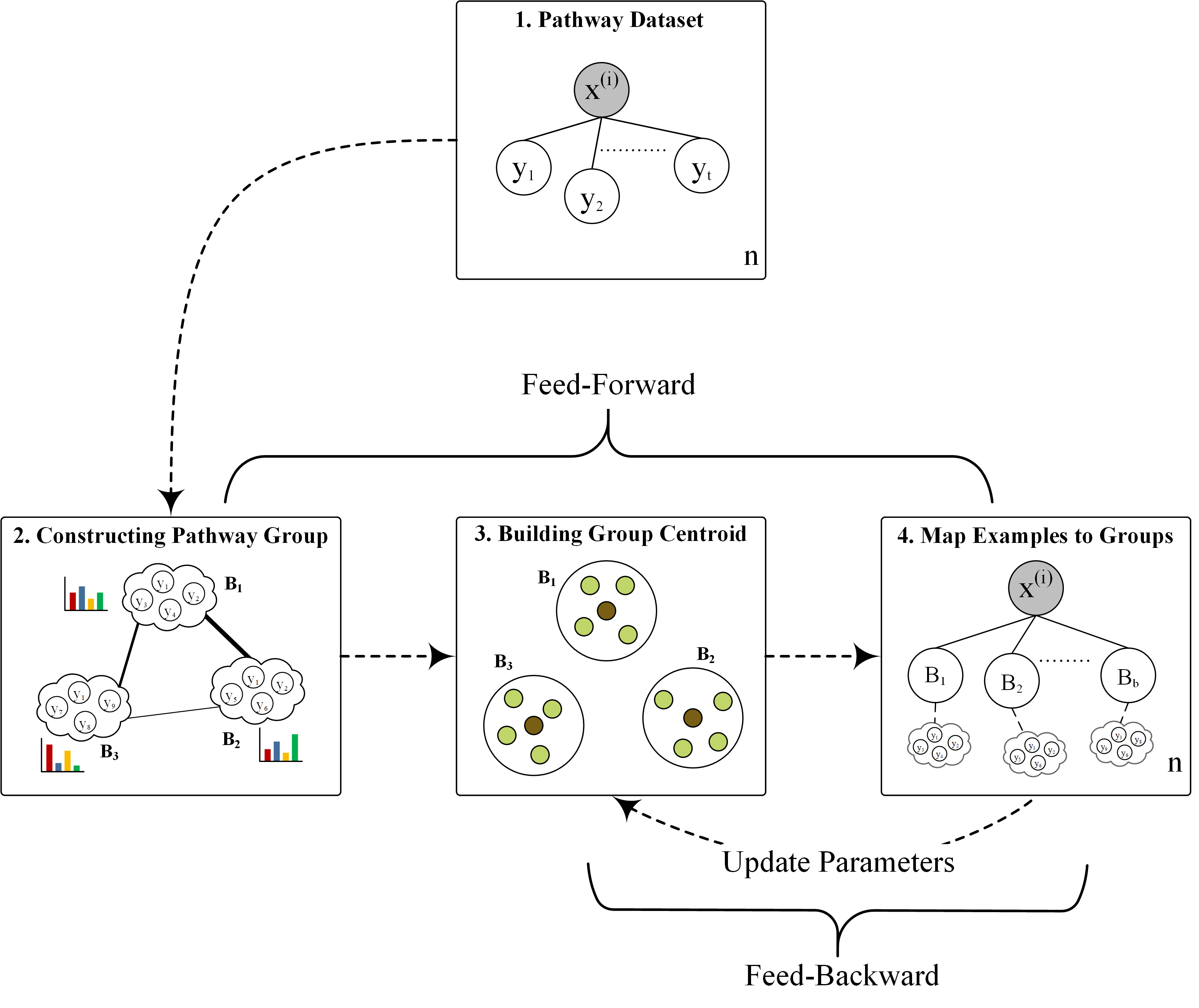
Relabeling metabolic pathway data with groups to improve prediction outcomes
Abdurrahman Abul-Basher and
Steven J. Hallam
ICCABS (to appear), 2021
code
This paper presents the reMap framework that performs mapping examples to a different set of labels, characterized as pathway groups, where a group comprises of statistically correlated pathways.
|
|
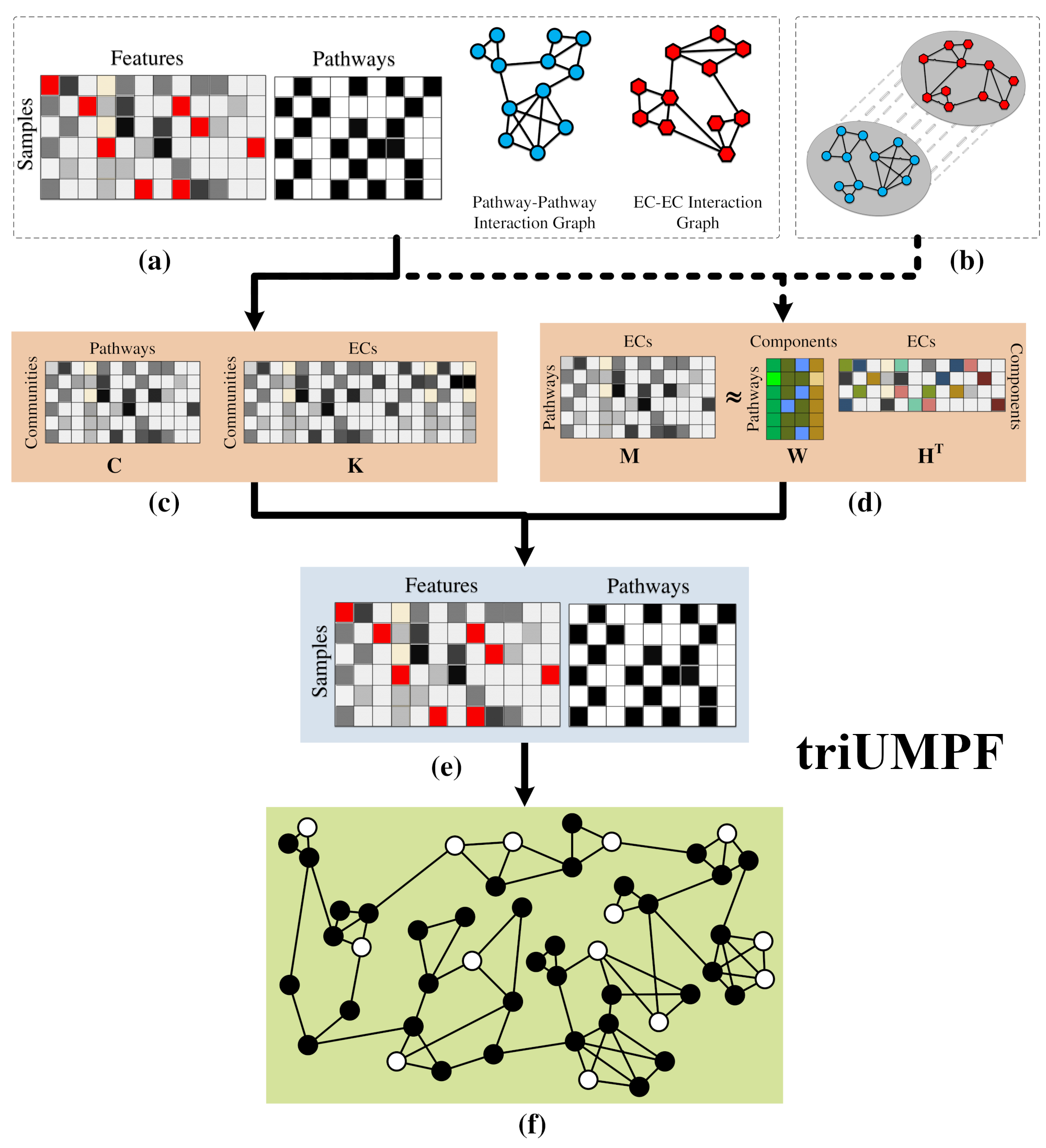
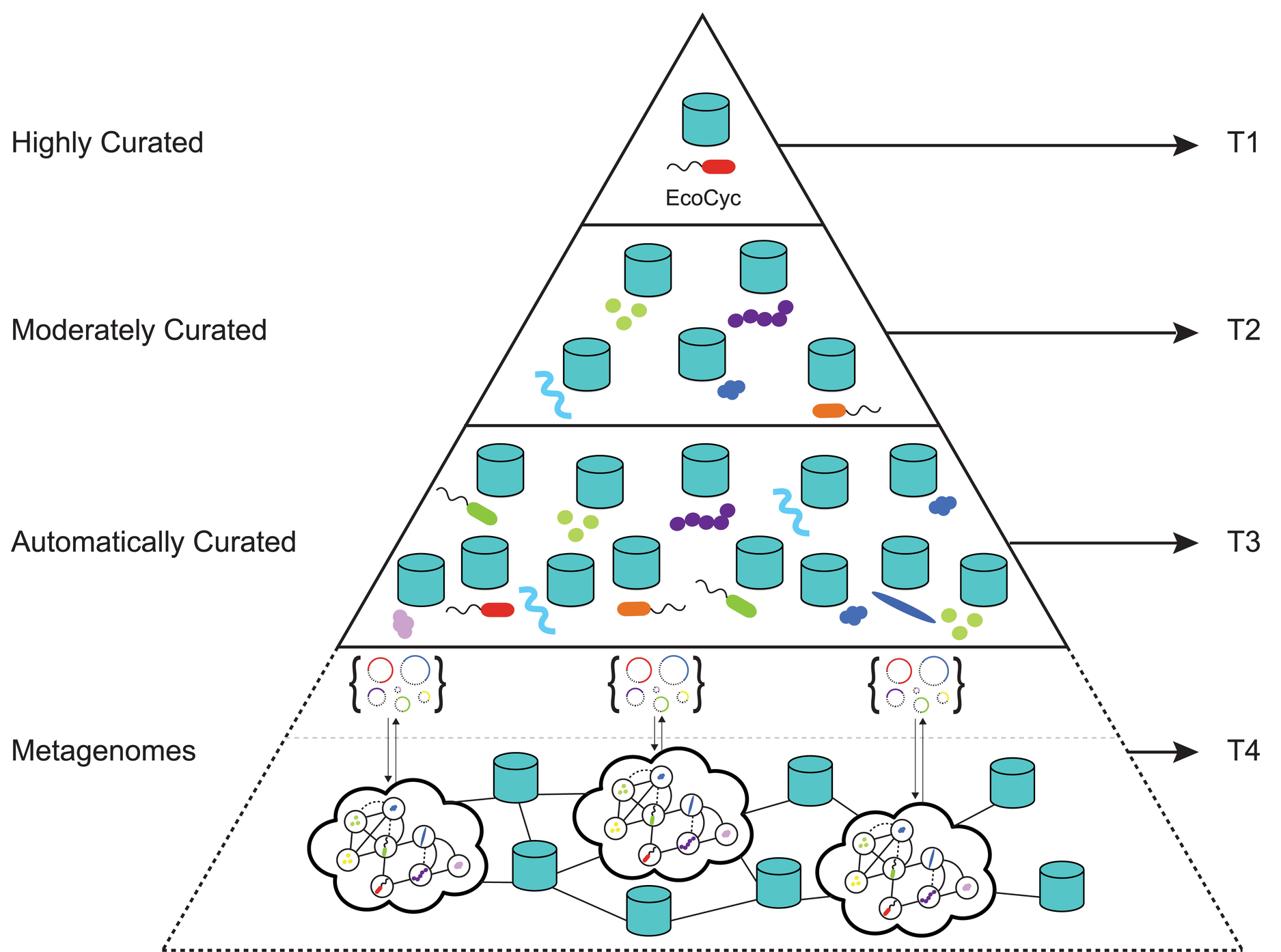
Metabolic pathway prediction using non-negative matrix factorization with improved precision
Abdurrahman Abul-Basher,
Ryan J.
McLaughlin, and
Steven J. Hallam
Journal of computational biology (JCB), 2021
code
This paper presents triUMPF (triple non-negative matrix factorization (NMF) with community
detection for metabolic pathway inference) that combines three stages of NMF to capture
myriad relationships between enzymes and pathways within a graph network followed by
community detection to extract higher order structure based on the clustering of vertices
sharing similar statistical properties.
|
|
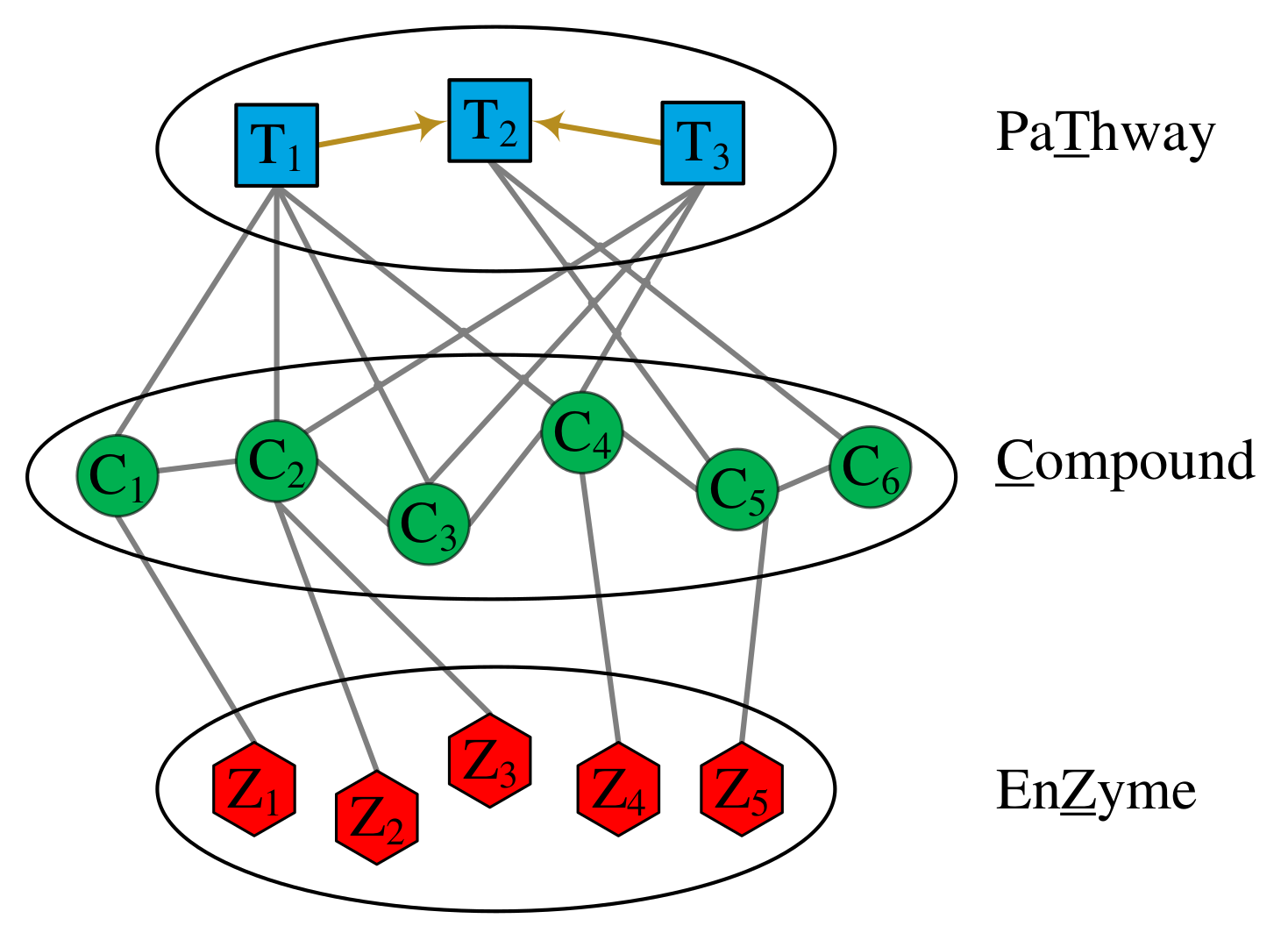
Leveraging heterogeneous network embedding for metabolic pathway prediction
Abdurrahman Abul-Basher and
Steven J. Hallam
Bioinformatics, 2020
code
This paper presents pathway2vec, a software package consisting of six representational
learning based modules used to automatically generate features for pathway inference.
|
|
Metabolic pathway inference using multi-label classification with rich pathway features
Abdurrahman Abul-Basher,
Ryan J.
McLaughlin, and
Steven J. Hallam
PLOS Computational Biology, 2020
code
This paper presents mlLGPR (multi-label logistic regression for pathway prediction)
framework that uses supervised multi-label classification and rich pathway features to infer
metabolic networks at the individual, population and community levels of organization.
|
|
|
|
|
|